Welcome to the PmiRtarbase 1.2
PmiRtarbase (a positive miRNA-target regulations database), is a database that curated experiment-supported evidence for microRNA (miRNA) and target gene associations. MicroRNAs (miRNAs) are ~22 nucleotide small non-coding RNA segments that are widely involved in the regulation of gene expression. Accumulating evidence shows that miRNAs not only inhibit the expression of targeted genes but also promote that of some targeted genes in specific conditions. Over the past decades, many miRNA-target databases have been developed from computational prediction and/or experimental validation perspectives. However, there is no database available to systematically collect positive miRNA-target associations that are essential in deciphering miRNA regulation mechanism. To promote the miRNA study, we developed a new database: PmiRtarbase that acquires validated positive miRNA-target interactions by mining published literature. It includes 312 curated associations between 119 miRNAs and 169 genes in 8 species from 130 studies and summarizes the conditions and detailed descriptions of the miRNA-target associations This elaborate database aims to serve as a beneficial resource for studying the miRNA positive regulation mechanism and miRNA-based therapeutics.
Current version
Release 1.2: March, 2022
Number of articles: 130
Number of species: 8
Number of target genes: 169
Number of miRNAs: 119
Number of miRNA-target interactions: 312
History:
March, 2022, the PmiRtarbase 1.2 was released.
May, 2020, the PmiRtarbase 1.1 was released.
Jan. 1, 2020, the original PmiRtarbase 1.0 was released.
Contact us
PmiRtarbase is free only for academic usage. For commercial usage, please Contact.
Dr. Peng Xu, Institute of computational science and technology, Guangzhou University, Guangzhou, Guangdong, 510006, China.
Email:
gdxupeng@gzhu.edu.cn
PmiRtarbase 1.0 Tutorial
MicroRNA (miRNA) is a class of non-coding single-stranded RNA molecules of approximately 22 nucleotides in length encoded by endogenous genes. They are involved in the regulation of post-transcriptional gene expression in plants and animals. For a long time in the past, many researchers believe that miRNAs only have negative regulatory relationships on genes, and have constructed multiple related databases. With the deepening of research, more and more positive regulatory relationships are discovered, which not only enriches and improves the regulatory mechanism of miRNAs, but also deepens researchers' understanding of it. However, there is currently no database that collects positive regulatory relationships. Based on this, we have constructed the PmiRtarbase database, a database of positive regulatory relationships between miRNAs and target genes.
PmiRtarbase (the Positive miRNA-target regulations database), is a database
that curated experiment-supported evidence for microRNA (miRNA) and target
gene associations.
We released version 1.0 of PmiRtarbase on January 1, 2020. All of the data are
from experimentally verified articles, a total of 110 articles. After collation,
267 miRNA-target records were obtained, which involved 8 species, 160 genes, and
98 miRNAs. The statistical information is shown in Table 1 and figure 1. In PmiRtarbase
version 1.0, users can check whether they have a positive regulatory relationship by
entering miRNA names and gene names, and export the query results. In addition, users
can also download all the regulatory relationship data in PmiRtarbase via the "Download"
button.
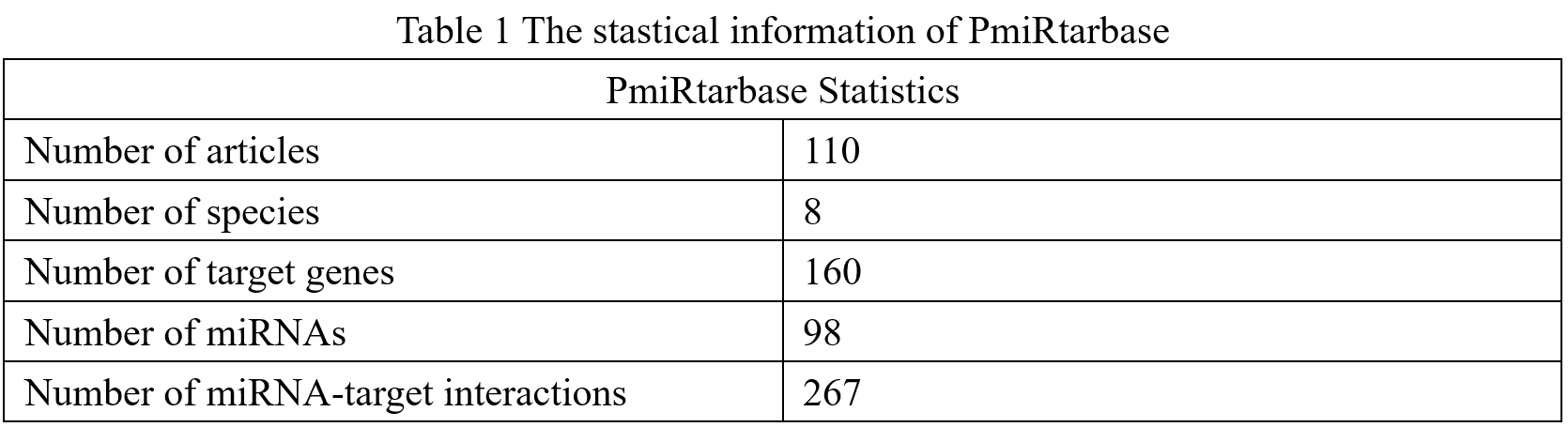
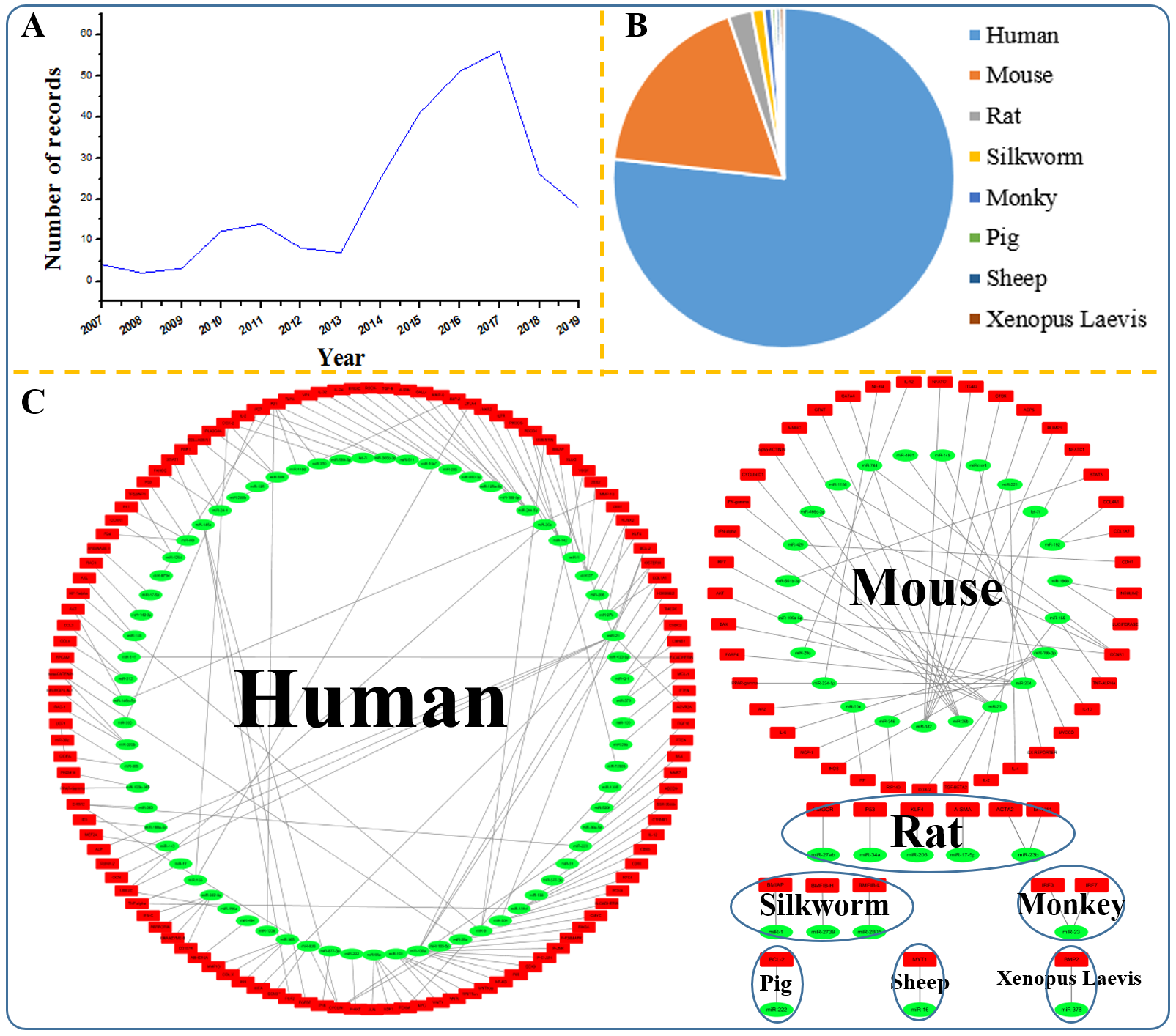
Figure 1 Visualization of PmiRtarbase statistics
The detailed usage of the database is as followings:
[1]:Visiting PmiRtarbase v1.0: Log on PmiRtarbase at http://www.lwb-lab.cn/PmiRtarbase/
[2]:Home page introduction. There are 4 modules in the "Home" section. Module 1 is a brief introduction to the database, module 2 is the statistical information of the database, module 3 is the historical version released, and module 4 is the contact information of the team members. See Figure 2 for details.
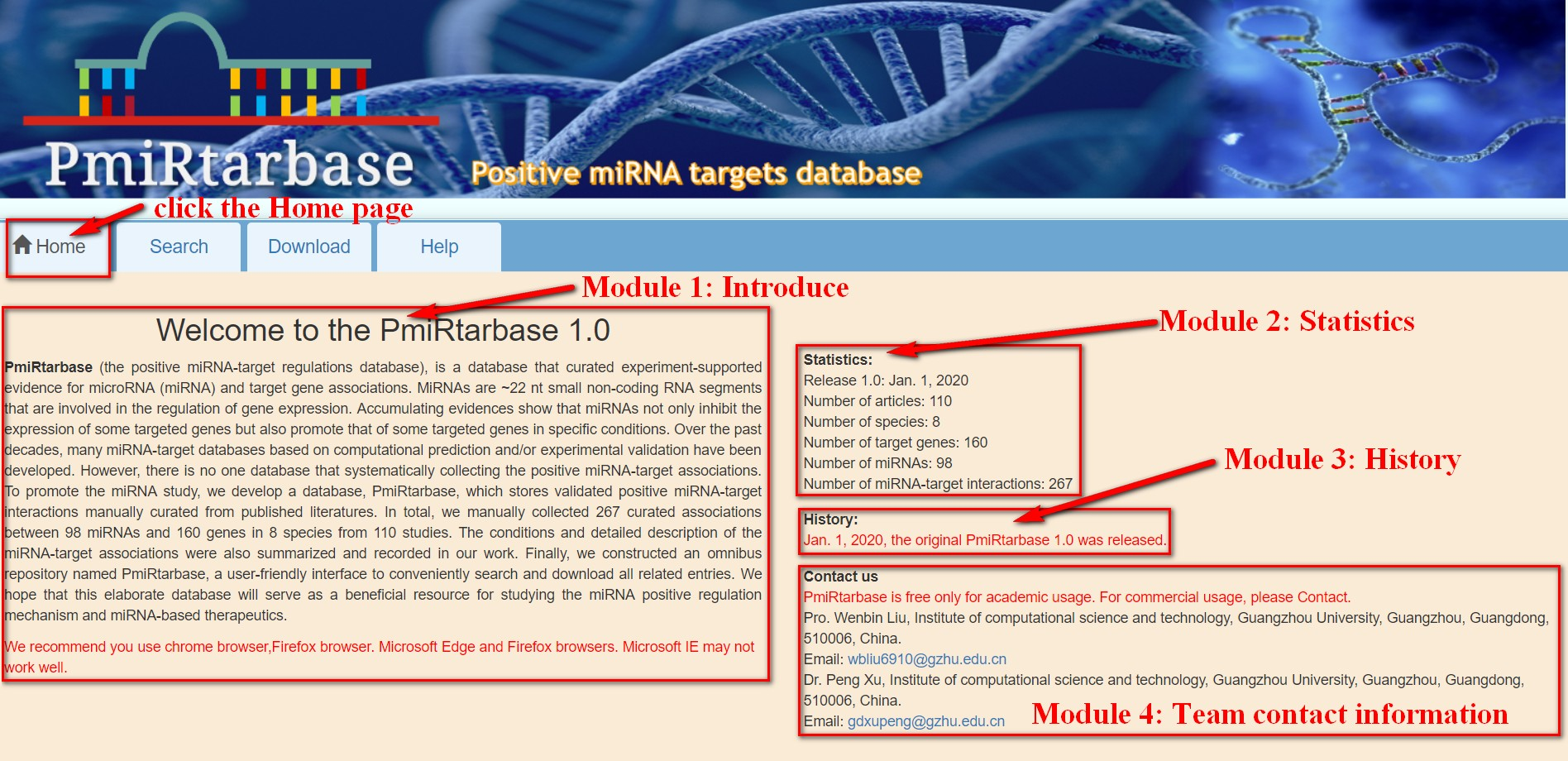
Figure 2 Home page introduction
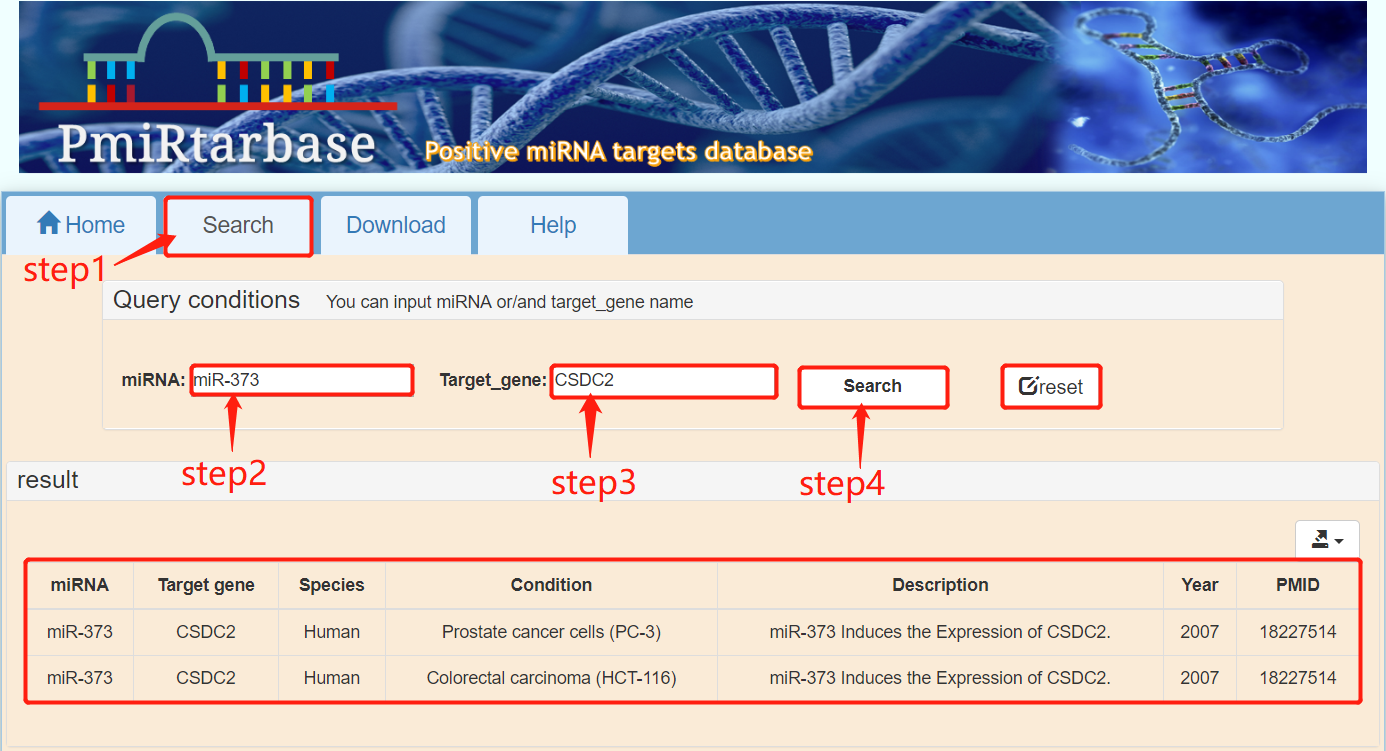
[3]:Query the positive regulatory relationship of miRNAs to genes. To search for the regulatory relationship of miRNAs to genes, click the "Search" button in the navigation bar. For example, you can search for the regulatory relationship between miRNA “miR-373” and gene “CSDC2” through the following steps. Step1: Click the "Search" button in the navigation bar; step2: Enter the miRNA name in the "miRNA" input box; step3: Enter the gene name in the "Target_gene" input box; step4: Click the "Search" button. Finally, the query results of the regulatory relationship between miR-373 and CSDC2 were obtained. The "reset" button in Figure 3 resets keywords.
[4]:How to enter keywords. In "Search", users can enter keywords in the following ways to get query results.
- Enter the miRNA and Target_gene names at the same time;
- Enter only miRNA name;
- Enter only the target gene name.
- miRNA-miRNA name
- Target gene-target gene name
- Species-the species to which the subject belongs
- Conditions-the diseased tissue of the experimental sample or the way the sample is processed
- Description-description of regulatory relationships in the literature
- Year-Year of Reference Publication
- pmid-the article number of the article in PubMed
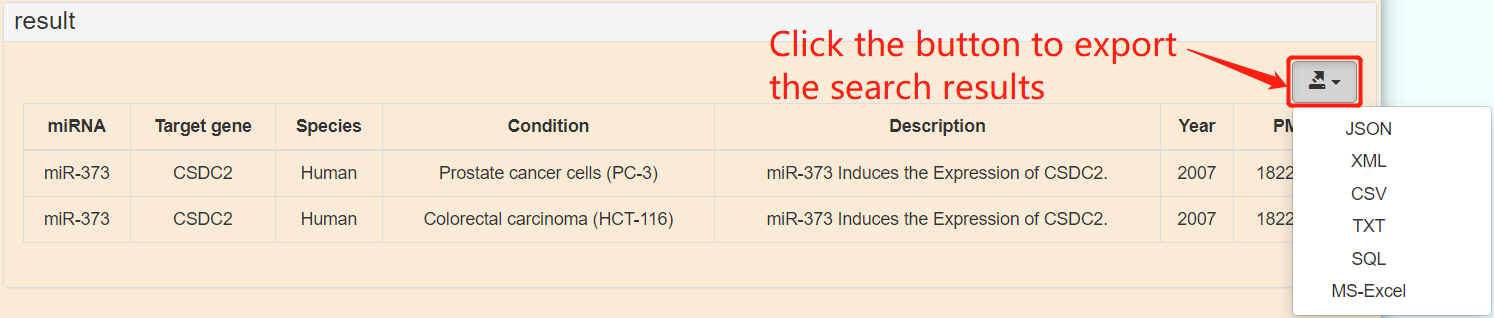
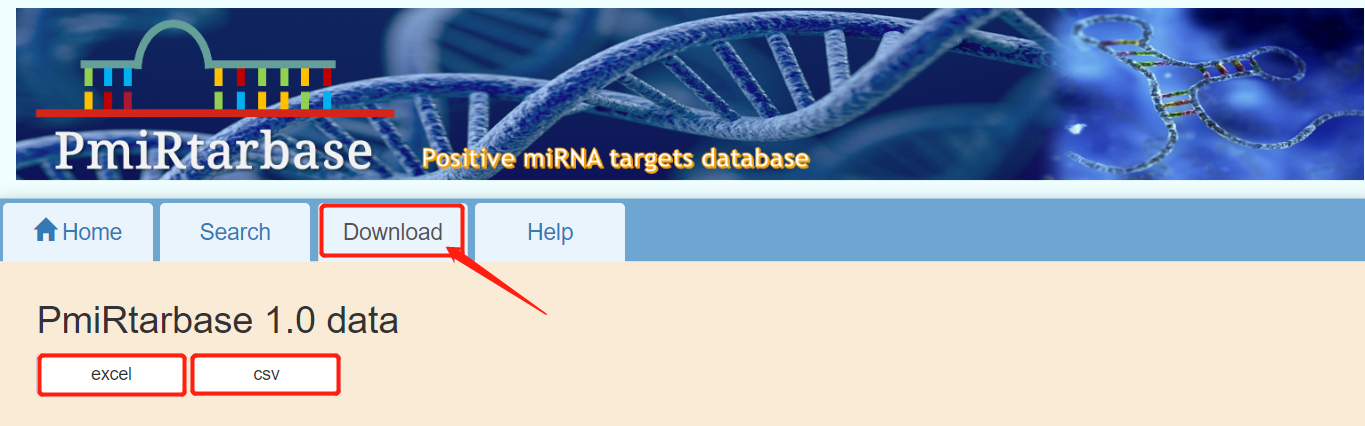
[7]:download. PmiRtarbase provides raw data download. If you need to download, you can click the "download" button in the navigation bar to download. Version 1.0 provides two data formats, excel and csv.
[8]: Currently, all data in PmiRtarbase is free only for academic users and please contact Dr. Peng Xu for commercial using. If you used the data in PmiRtarbase 1.0, please cite….
